Protein interaction and complex modeling
Protein interaction networks are central to any understanding of cellular processes, and though many thousands are now known, few initiatives to uncover them pay much attention to one of the best sources of data available: complexes of known 3D structure. We thus study protein interactions by considering known 3D structures. We use 3D complexes to interrogate interactions identified by other methods (e.g. yeast two-hybrids) and to predict specific interactions within protein families. A major initiative in the group is related building as complete models as possible for all interacting proteins and complexes in a whole cell (see Figure). This is particularly useful when combined with experimental methods like electron microscopy, cryo-electron tomography, mass-spectrometry or proteome-scale interaction discovery. We have also participated in attempts to decipher the entire interacting proteome within organisms.
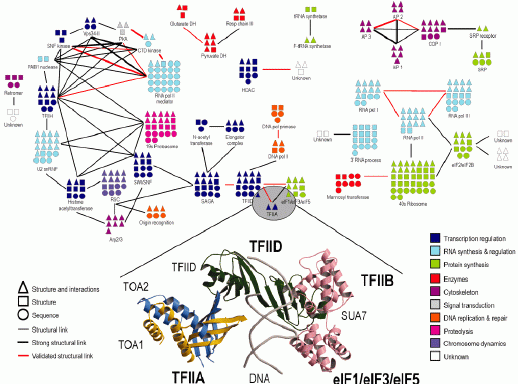
Network depicting modellable complexes in Yeast (see Aloy et al, Science 2004).
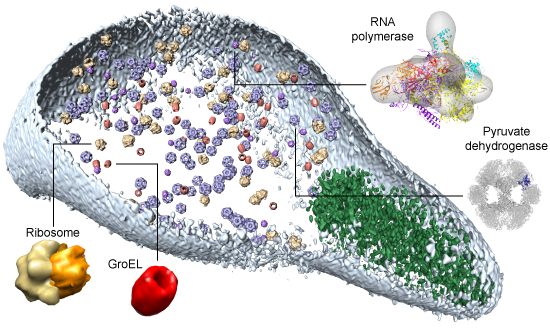
Fitting modelled complexes into a tomogram of Mycoplasma pneumoniae (see Kuehner et al, Science 2009).
- An organelle-specific protein landscape identifies novel diseases and molecular mechanisms
Boldt, van Reeuwijk J, Lu Q, Koutroumpas et al Nat Commun 2016 - Proteome organization in a genome-reduced bacterium.
Kühner et al. Science 2009 - Structural systems biology: modelling protein interactions.
Aloy & Russell. Nat Rev Mol Cell Biol 2006 - Proteome survey reveals modularity of the yeast cell machinery.
Gavin et al. Nature 2006 - Structure-based assembly of protein complexes in yeast.
Aloy et al. Science 2004



