Protein-peptide interactions & motifs
A major current challenge in biology is to discover and understand short protein tretches that mediate functional interactions. We have developed several methods for predicting these kinds of interactions, and continue to apply them to new problems in biology.
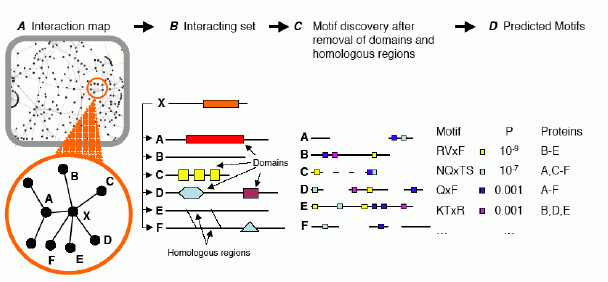
Schematic outlining DiLiMOT: our appraoch for finding protein linear motifs that mediate protein-protein interactions. See Neduva et al, PLoS Biol. 2005).
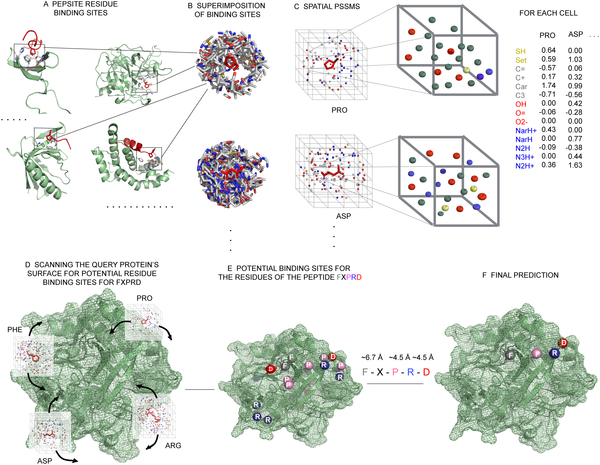
Schematic detailing PepSite: our approach for finding peptide binding sites on protein surfaces. See Petsalaki et al, PLoS Comp. Biol 2009)
- EZHIP/CXorf67 mimics K27M mutated oncohistones and functions as an intrinsic inhibitor of PRC2 function in aggressive posterior fossa ependymoma
Huebner et al Neuro Oncol 2019 Jul 11;21(7):878-889. - PepSite: prediction of peptide-binding sites from protein surfaces.
Trabuco et al Nucleic Acids Res 2012 40 W423-W427 - Accurate prediction of peptide binding sites on protein surfaces.
Petsalaki et al. PLoS Comput Biol. 2009 Mar;5(3):e1000335. Epub 2009 Mar 27. - Peptide-mediated interactions in biological systems: new discoveries and applications.
Petsalaki & Russell. Curr Opin Biotechnol. 2008 Aug;19(4):344-50. Epub 2008 Jul 12. - Systematic discovery of in vivo phosphorylation networks.
Linding et al. Cell. 2007 Jun 29;129(7):1415-26. Epub 2007 Jun 14. - Systematic discovery of new recognition peptides mediating protein interaction networks.
Neduva et al. PLoS Biol. 2005 Dec;3(12):e405. Epub 2005 Nov 15.



