The impact of mutations and post-translational modifications on biomolecular interactions
High-throughput sequencing (HTS) and proteomics approaches now mean that tens of thousands of positional variations within genes or proteins can be identified in a single experiment. The vast majority of such variations are still new and require advanced methods to identify those that are important for disease or biological mechanism. We develop methods and perform analyses to interrogate these data. This work builds on our 15 years experience in relating structures to the interactome and to date we have studied dozens of disease mutation and post-translational modification datasets. Our aim is, for each mutation/modification and for each gene/protein to assess the overall impact on biomolecular interactions with sub-aims to help identify biological switches, deleterious or oncogenic mutations and other insights arising from a more mechanistic understanding of such changes in proteins. We continue to develope our Mechismo server to help scientists interpret these large and complicated datasets.

Network showing predicted functional mutations in Pancreatic cancer (see Betts et al Nucl Acids Res 2015).
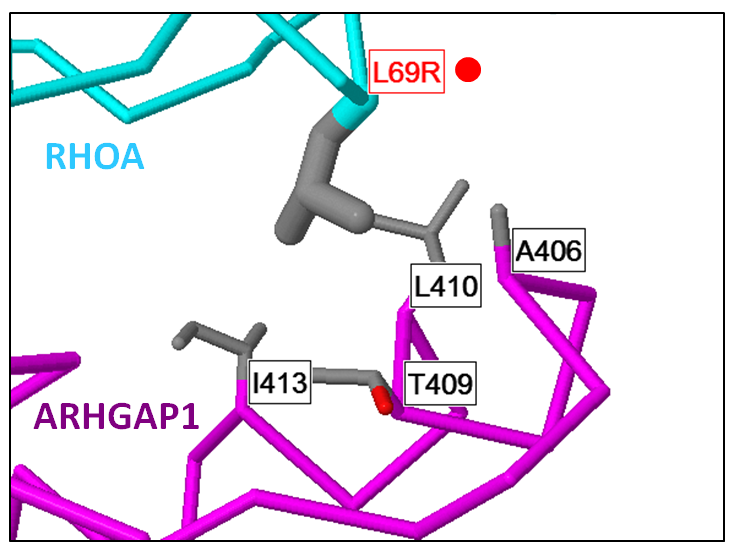
Predicted destablizing RhoA/ArhGap1 mutation in Lymphoma (see Betts et al Nucl Acids Res 2015).
- Systematic identification of phosphorylation-mediated protein interaction switches
Betts et al PLoS Comp Biol 2016 - Insights into cancer severity from biomolecular interaction mechanisms
Raimondi, Singh et al Sci. Rep. 2016 - An organelle-specific protein landscape identifies novel diseases and molecular mechanisms
Boldt, van Reeuwijk J, Lu Q, Koutroumpas et al Nat Commun 2016 - Mechismo: predicting the mechanistic impact of mutations and modifications on molecular interactions.
Betts et al. Nucleic Acids Res 2015 - Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing.
Richter et al. Nat Genet 2012



